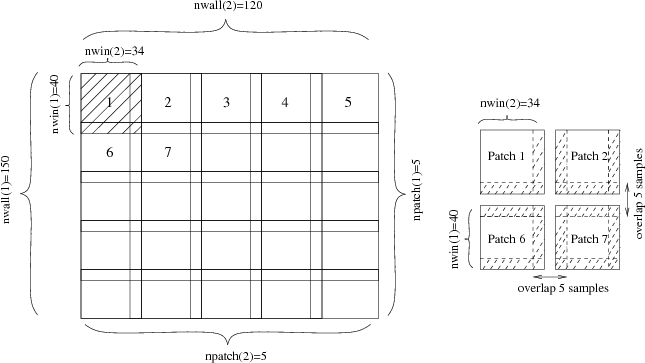
sfpatch breaks the input data into local windows or “patches”, possibly with overlap.
The patching technique is explained by Jon Claerbout in Nonstationarity: patching chapter from Image Estimation by Example.
Suppose you have a 1-D signal with 10 samples:
You can divide it, for example, into two patches with 5 samples each:
or into 5 overlapping patches with 3 samples each:
If you specify only the patch size (w= parameter), sfpatch tries to select the number of patches to achieve a good overlap:
If you put overlapped patches back together, their amplitudes add in the overlapped regions:
bash$ < data.rsf sfdd type=int | sfdisfil
0: 0 1 2 3 4 5 6 7 8 9
bash$< patch.rsf sfpatch inv=y | sfdd type=int | sfdisfil
0: 0 2 6 6 8 10 12 14 8 9
unless you use the weight option:
bash$ sfpatch easily handles multidimensional data: w= and p= become lists of dimensions, and the number of dimensions in the output effectively doubles. sfpatch is useful in two applications:
- crude handling of non-stationarity when processing non-stationary signals with locally stationary filters,
- making non-parallel tasks data-parallel.
A simple example from rsf/rsf/mona illustrates the second use.


Anisotropic diffusion, implemented by sfimpl2, is an effective but relatively slow process. By breaking the input 512×512 image into nine 200×200 overlapping patches and by processing them in parallel on a multi-core computer, we can achieve a significant speed-up without rewriting the original program.
 Sometimes it is convenient to have a quick access to all results in your experiment.
Sometimes it is convenient to have a quick access to all results in your experiment.