|
|
|
|
Homework 6 |
Your task: Change the program for reverse-time migration to implement forward-time modeling using the ``exploding reflector'' approach.
cd hw6/hyper
scons viewto generate figures and display them on your screen.
scons wave.vplto observe a movie of reverse-time wave extrapolation.
scons viewagain to observe the differences.
|
|---|
|
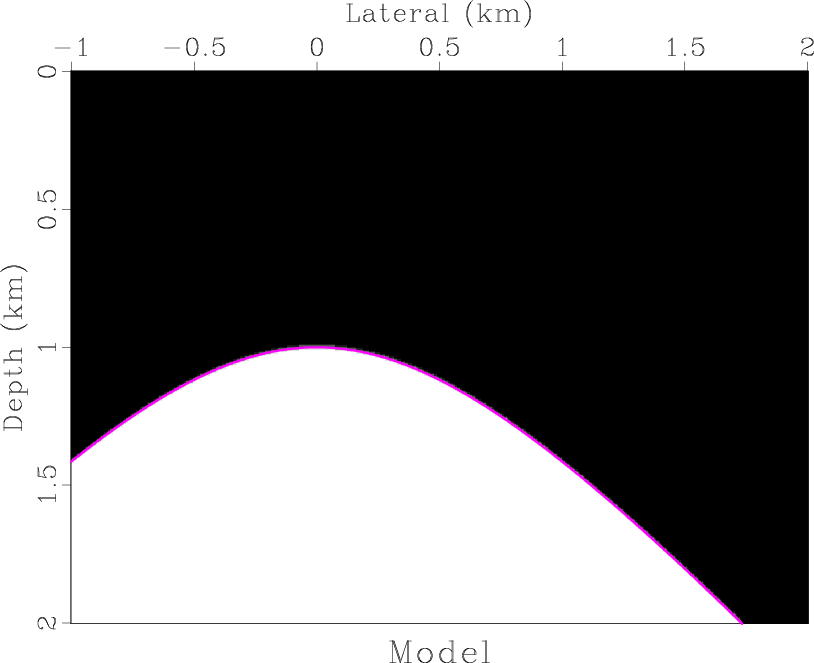
model
Figure 1. Synthetic velocity model with a hyperbolic reflector. |
|
|


|
|---|
|
data,image
Figure 2. (a) Synthetic zero-offset data corresponding to the model in Figure 1. (b) Image generated by reverse-time exploding-reflector migration. The location of the exact reflector is indicated by a curve. |
|
|
from rsf.proj import *
# Make a reflector model
Flow('refl',None,
'''
math n1=10001 o1=-4 d1=0.001 output="sqrt(1+x1*x1)"
''')
Flow('model','refl',
'''
window min1=-3 max1=3 j1=5 |
unif2 n1=401 d1=0.005 v00=1,2 |
put label1=Depth unit1=km label2=Lateral unit2=km |
smooth rect1=3
''')
Plot('model',
'''
window min2=-1 max2=2 |
grey allpos=y bias=1 title=Model
''')
Plot('refl',
'''
graph wanttitle=n wantaxis=n yreverse=y
min1=-1 max1=2 min2=0 max2=2
plotfat=3 plotcol=4
''')
Result('model','model refl','Overlay')
# Kirchoff zero-offset modeling
Flow('dip','refl','math output="x1/input" ')
Flow('data','refl dip',
'''
kirmod nt=1501 dt=0.004 freq=10
ns=1201 s0=-3 ds=0.005 nh=1 dh=0.01 h0=0
twod=y vel=1 dip=${SOURCES[1]} verb=y |
window | costaper nw2=200
''')
Result('data',
'''
window min2=-2 max2=2 min1=1 max1=4 |
grey title="Zero-Offset Data" grid=y
label1=Time unit1=s label2=Distance unit2=km
''')
# Reverse-time migration
proj = Project()
prog = proj.Program('rtm.c')
Flow('image wave','model %s data' % prog[0],
'''
pad beg1=200 end1=200 | math output=0.5 |
./${SOURCES[1]} data=${SOURCES[2]}
wave=${TARGETS[1]} jt=20 n0=200
''')
# Display wave propagation movie
Plot('wave',
'''
window min2=-1 max2=2 min1=0 max1=2 f3=33 |
grey wanttitle=n gainpanel=all
''',view=1)
# Display image
Plot('image',
'''
window min2=-1 max2=2 min1=0 max1=2 |
grey title=Image
''')
Result('image','image refl','Overlay')
End()
|
/* 2-D zero-offset reverse-time migration */
#include <rsf.h>
#ifdef _OPENMP
#include <omp.h>
#endif
static int n1, n2;
static float c0, c11, c21, c12, c22;
static void laplacian(float **uin /* [n2][n1] */,
float **uout /* [n2][n1] */)
/* Laplacian operator, 4th-order finite-difference */
{
int i1, i2;
#ifdef _OPENMP
#pragma omp parallel for private(i2,i1) shared(n2,n1,uout,uin,c11,c12,c21,c22,c0)
#endif
for (i2=2; i2 < n2-2; i2++) {
for (i1=2; i1 < n1-2; i1++) {
uout[i2][i1] =
c11*(uin[i2][i1-1]+uin[i2][i1+1]) +
c12*(uin[i2][i1-2]+uin[i2][i1+2]) +
c21*(uin[i2-1][i1]+uin[i2+1][i1]) +
c22*(uin[i2-2][i1]+uin[i2+2][i1]) +
c0*uin[i2][i1];
}
}
}
int main(int argc, char* argv[])
{
int it,i1,i2; /* index variables */
int nt,n12,n0,nx, jt;
float dt,dx,dz, dt2,d1,d2;
float **vv, **dd;
float **u0,**u1,u2,**ud; /* tmp arrays */
sf_file data, imag, modl, wave; /* I/O files */
/* initialize Madagascar */
sf_init(argc,argv);
/* initialize OpenMP support */
omp_init();
/* setup I/O files */
modl = sf_input ("in"); /* velocity model */
imag = sf_output("out"); /* output image */
data = sf_input ("data"); /* seismic data */
wave = sf_output("wave"); /* wavefield */
/* Dimensions */
if (!sf_histint(modl,"n1",&n1)) sf_error("n1");
if (!sf_histint(modl,"n2",&n2)) sf_error("n2");
if (!sf_histfloat(modl,"d1",&dz)) sf_error("d1");
if (!sf_histfloat(modl,"d2",&dx)) sf_error("d2");
if (!sf_histint (data,"n1",&nt)) sf_error("n1");
if (!sf_histfloat(data,"d1",&dt)) sf_error("d1");
if (!sf_histint(data,"n2",&nx) || nx != n2)
sf_error("Need n2=%d in data",n2);
n12 = n1*n2;
if (!sf_getint("n0",&n0)) n0=0;
/* surface */
if (!sf_getint("jt",&jt)) jt=1;
/* time interval */
sf_putint(wave,"n3",1+(nt-1)/jt);
sf_putfloat(wave,"d3",-jt*dt);
sf_putfloat(wave,"o3",(nt-1)*dt);
dt2 = dt*dt;
/* set Laplacian coefficients */
d1 = 1.0/(dz*dz);
d2 = 1.0/(dx*dx);
c11 = 4.0*d1/3.0;
c12= -d1/12.0;
c21 = 4.0*d2/3.0;
c22= -d2/12.0;
c0 = -2.0 * (c11+c12+c21+c22);
/* read data and velocity */
dd = sf_floatalloc2(nt,n2);
sf_floatread(dd[0],nt*n2,data);
vv = sf_floatalloc2(n1,n2);
sf_floatread(vv[0],n12,modl);
/* allocate temporary arrays */
u0=sf_floatalloc2(n1,n2);
u1=sf_floatalloc2(n1,n2);
ud=sf_floatalloc2(n1,n2);
for (i2=0; i2<n2; i2++) {
for (i1=0; i1<n1; i1++) {
u0[i2][i1]=0.0;
u1[i2][i1]=0.0;
ud[i2][i1]=0.0;
vv[i2][i1] *= vv[i2][i1]*dt2;
}
}
/* Time loop */
for (it=nt-1; it >= 0; it-) {
sf_warning("%d;",it);
laplacian(u1,ud);
#ifdef _OPENMP
#pragma omp parallel for private(i2,i1,u2) shared(ud,vv,it,u1,u0,dd,n0)
#endif
for (i2=0; i2<n2; i2++) {
for (i1=0; i1<n1; i1++) {
/* scale by velocity */
ud[i2][i1] *= vv[i2][i1];
/* time step */
u2 =
2*u1[i2][i1]
- u0[i2][i1]
+ ud[i2][i1];
u0[i2][i1] = u1[i2][i1];
u1[i2][i1] = u2;
}
/* inject data */
u1[i2][n0] += dd[i2][it];
}
if (0 == it%jt)
sf_floatwrite(u1[0],n12,wave);
}
sf_warning(".");
/* output image */
sf_floatwrite(u1[0],n12,imag);
exit (0);
}
|


|
|---|
|
zvel,zref
Figure 3. (a) Sigsbee velocity model. (b) Approximate reflectivity of the Sigsbee model (an ideal image). |
|
|
Your task: Apply your exploding-reflector modeling and migration program from the previous task to generate zero-offset data for Sigsbee and image it.
cd hw6/sigsbee
scons viewto generate figures and display them on your screen.
from rsf.proj import *
# Download velocity model from the data server
##############################################
vstr = 'sigsbee2a_stratigraphy.sgy'
Fetch(vstr,'sigsbee')
Flow('zvstr',vstr,'segyread read=data')
Flow('zvel','zvstr',
'''
put d1=0.025 o2=10.025 d2=0.025
label1=Depth unit1=kft label2=Lateral unit2=kft |
scale dscale=0.001
''')
Result('zvel',
'''
window f1=200 |
grey title=Velocity titlesz=7 color=j
screenratio=0.3125 screenht=4 labelsz=5
mean=y
''')
# Compute approximate reflectivity
##################################
Flow('zref','zvel',
'''
depth2time velocity=$SOURCE nt=2501 dt=0.004 |
ai2refl | ricker1 frequency=10 |
time2depth velocity=$SOURCE
''')
Result('zref',
'''
window f1=200 |
grey title=Reflectivity titlesz=7
screenratio=0.3125 screenht=4 labelsz=5
''')
End()
|
|
map2
Figure 4. Map of the Blake Outer Ridge area. An region of known gas hydrate distribution as mapped from seismic bottom simulating reflectors (BSR) is highlighted. The seismic survey area is marked by a rectangle. |

|
|---|---|
|
|
The following figures show the dataset at different stages of seismic data processing: from initial data to an image in depth.
Your task: Modify the data processing sequence to create a justifiably better image.

|
|---|
|
nmo
Figure 5. Common midpoint gather (left), velocity analysis panel using normal moveout (middle), and common midpoint gather after normal moveout (right). A curve in the middle plot indicates an automatically picked velocity trend. |
|
|

|
|---|
|
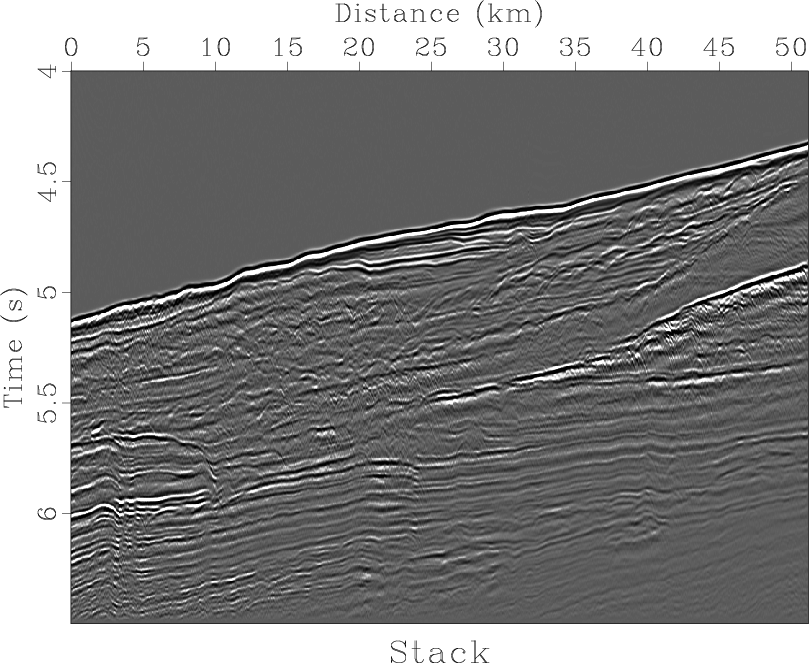
noff,stack
Figure 6. (a) Near-offset section. (b) Normal moveout stack. |
|
|


|
|---|
|
picks,vel
Figure 7. (a) Picked stacking velocity. (b) Interval velocity estimated by Dix inversion. |
|
|

|
|---|
|
image
Figure 8. Seismic image created by converting stacked data from time to depth. Can you identify geological features that are not properly imaged? |
|
|
from rsf.proj import *
# get data
Fetch('cmps-tp.HH','blake')
# CMP (common midpoint) gathers
Flow('cmps','cmps-tp.HH',
'dd form=native | reverse which=2')
# one CMP
Flow('cmp','cmps',
'''
window f3=950 n3=1 max1=6 |
put o2=0.0 d2=1
''')
Plot('cmp',
'''
grey title="CMP gather"
unit1=s label2=Offset unit2=km
''')
# Near-offset section
Result('noff','cmps',
'''
window n2=1 n3=1024 |
grey title="Near Offset Section"
unit1=s label2=Distance unit2=km
''')
# offset maps
Flow('off1',None,'math n1=24 o1=0.4 d1=0.1 output=x1')
Flow('off2',None,'math n1=24 o1=2.8 d1=0.05 output=x1')
Flow('off','off1 off2','cat axis=1 ${SOURCES[1]}')
Flow('offs','off','spray n=111')
# Velocity analysis
###################
vscan = '''
vscan offset=${SOURCES[1]}
v0=1.4 nv=61 dv=0.005 half=n semblance=y
'''
pick = 'pick rect1=20 rect2=3 vel0=1.5'
# compute semblance for one CMP
Flow('vscan','cmp off',vscan)
Plot('vscan',
'''
grey color=j allpos=y title="Velocity Scan"
unit1=s label2=Velocity unit2=km/s pclip=100
''')
# pick maximum semblance for one CMP
Flow('pick','vscan',pick)
Plot('pick0','pick',
'''
graph transp=y yreverse=y min2=1.4 max2=1.7
plotcol=7 plotfat=10 pad=n wanttitle=n wantaxis=n
''')
Plot('pick1','pick',
'''
graph transp=y yreverse=y min2=1.4 max2=1.7
plotcol=0 plotfat=1 pad=n wanttitle=n wantaxis=n
''')
Plot('vscan2','vscan pick0 pick1','Overlay')
# compute semblance for every 10th CMP
Flow('vscans','cmps offs','window j3=10 | ' + vscan)
# pick max semblance for every 10th CMP
Flow('picks0','vscans',pick)
# interpolate picks on the original grid
Flow('picks','picks0',
'transp | remap1 n1=1105 d1=0.05 o1=0 | transp')
Result('picks',
'''
grey color=j scalebar=y barreverse=y
allpos=y bias=1.4
title="Stacking Velocity"
label1=Time unit1=s label2=Distance unit2=km
''')
# Normal moveout and stack
##########################
nmo = '''
nmo offset=${SOURCES[1]}
velocity=${SOURCES[2]} half=n
'''
Flow('nmo','cmp off pick',nmo)
Plot('nmo',
'''
grey title="Normal Moveout"
unit1=s label2=Offset unit2=km
''')
Result('nmo','cmp vscan2 nmo','SideBySideAniso')
Flow('nmos','cmps off picks',nmo)
Flow('stack','nmos','stack')
Result('stack',
'''
window n2=1024 |
grey title=Stack
unit1=s label2=Distance unit2=km
''')
# Convert from time to depth
############################
# Dix inversion
Flow('semb','vscans picks0','slice pick=${SOURCES[1]}')
Flow('vel0','picks0 semb',
'dix rect1=20 rect2=2 weight=${SOURCES[1]}')
# interpolate on the original grid
Flow('vel','vel0',
'transp | remap1 n1=1105 d1=0.05 o1=0 | transp')
Result('vel',
'''
grey color=j scalebar=y barreverse=y
allpos=y bias=1.4
title="Interval Velocity"
label1=Time unit1=s label2=Distance unit2=km
''')
Flow('image','stack vel',
'''
time2depth velocity=${SOURCES[1]}
intime=y dz=0.005 nz=1001
''')
Result('image',
'''
window n2=1024 min1=3 | grey title=Image
label1=Depth unit1=km label2=Distance unit2=km
''')
End()
|
cd hw6/blake
scons viewto generate figures and display them on your screen.
scons viewagain.
|
|
|
|
Homework 6 |