|
|
|
|
2-D Seismic Data Processing Exercise |
The next step is to apply NMO-based velocity analysis to the Nankai dataset (Keys and Foster, 1998).
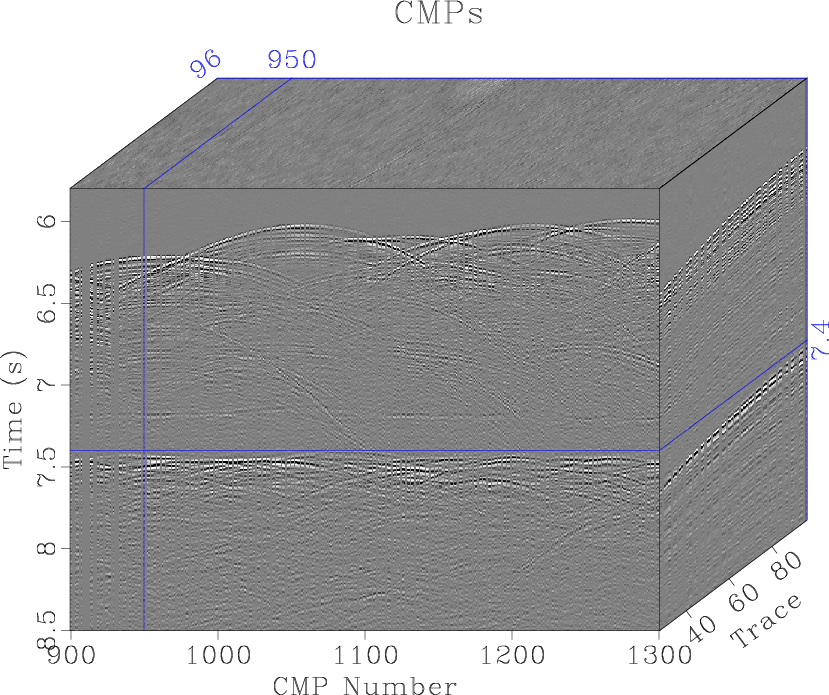
scons cmps.viewto display the input data (Figure 4.)
Hint: read the documentation for sfpow by running
sfpowwithout arguments or by checking the ``program of the month'' blog post
|
|---|
|
cmps
Figure 4. Nankai dataset after preprocessing and sorting into CMP gathers. |
|
|
scons cmp1.viewto display the selected gather (Figure 5.)

|
|---|
|
cmp1
Figure 5. CMP gather selected for velocity analysis. |
|
|
Run
scons vscan.view scons nmo.viewto observe the result of velocity analysis on the selected CMP gather using automatic picking (Figure 6.)


|
|---|
|
vscan,nmo1
Figure 6. NMO velocity analysis with automatic picking applied to the selected CMP gather. |
|
|
Run
scons picks.viewto run the semblance analysis with automatic picking on every CMP gather. The result is shown in Figure 7.

|
|---|
|
picks
Figure 7. NMO stacking velocity picked automatically from the entire line. |
|
|
Run
scons nmos.view scons stack.viewto display the result (Figures 8 and 9.)

|
|---|
|
nmos
Figure 8. Nankai dataset after normal-moveout correction. |
|
|

|
|---|
|
stack
Figure 9. NMO stack of the Nankai dataset. |
|
|
|
|
|
|
2-D Seismic Data Processing Exercise |